|
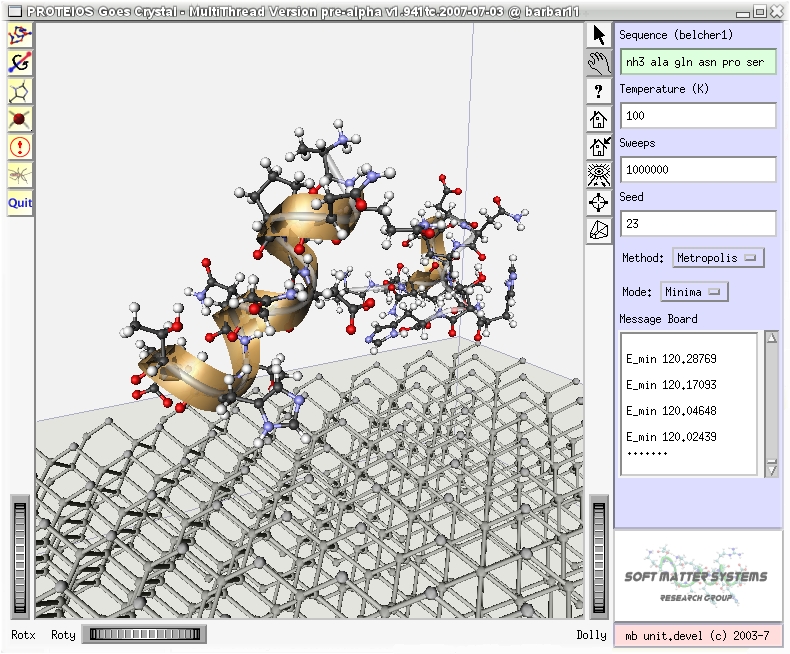
Monte Carlo simulation package for folding, aggregation, and adsorption of
small proteins
|

|
Two implicit-solvent protein force fields incorporated
|

|
Metal and semiconductor substrate materials with arbitrary surface orientations
|

|
Monte Carlo methods: Metropolis, multicanonical
sampling, multi-core multicanonical sampling, multi-core parallel tempering, Wang-Landau sampling
|

|
Simple minimization algorithms: simulated annealing, two variants of energy-landscape paving
|

|
Visualization interface based on the OpenInventor™ graphics library; current
implementation is based on Coin3D™; quadbuffer stereo
mode included, i.e., NVIDIA® 3D Vision™ capable
|